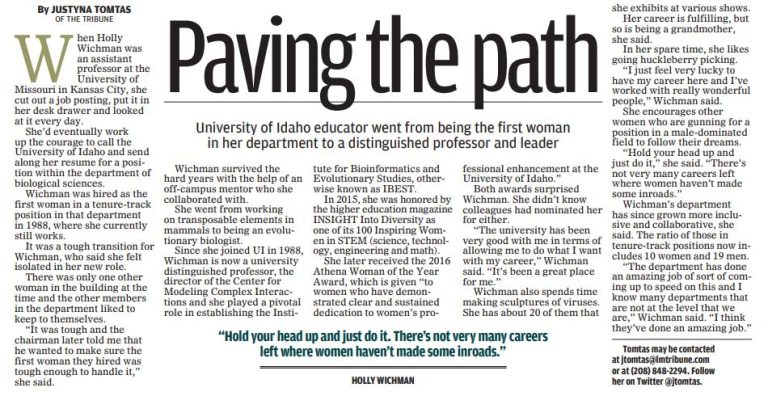
CMCI Participant Award Winners
Spring means we’re nearing the end of the school year – and various award banquets and receptions. We’re proud that so many CMCI participants received awards this year. 2019 College of Science Staff, Faculty, and Graduate Student Awards Nominations for the staff and faculty awards were made by members of the College of Science executive…