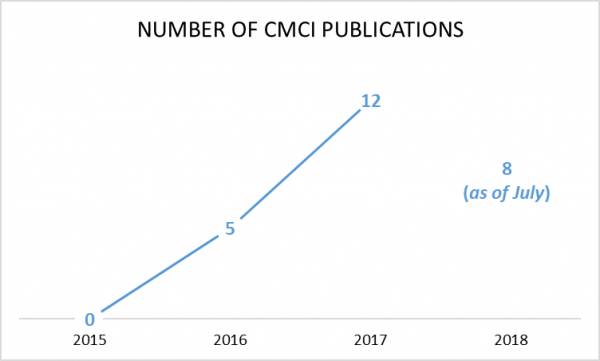
Since our inception in 2015, CMCI has seen an increase each year in the number of articles published.
To view a full list of titles published with links to the full articles, click here.

Since our inception in 2015, CMCI has seen an increase each year in the number of articles published.
To view a full list of titles published with links to the full articles, click here.
The Friday Letter is the university’s weekly update directly from the president. Today several CMCI faculty were mentioned:
U of I researchers are trying to answer questions that could help slow the spread of antibiotic drug resistance, develop techniques to produce vaccines more quickly and help with treatment of malaria thanks to three federal grants from the National Institutes of Health (NIH). The awards to Eva Top, Craig Miller and Holly Wichman in the College of Science and Shirley Luckhart in the colleges of Science and Agricultural and Life Sciences, total nearly $5 million to support multiyear studies aimed at improving disease treatment.
We love it when CMCI research news reaches the general public!

Nearly 35 researchers and scientists from the University of Idaho (Moscow, Idaho), Brown University (Providence, Rhode Island) and the University of Vermont (Burlington, Vermont) recently gathered for their first in-person “All-Hands Meeting” for the GenoPheno EPSCoR Track-2 award of $6M. The 3-day session hosted by CMCI was full of brainstorming, collaboration and team-building activities.
Project Team: Craig Miller (PI), Aniruddha Belsare, JT Van Leuven
Start Date: June 2018
Context
Rabies kills an estimated 59,000 people every year. Most of these deaths are from the poorest sectors of society in low- and middle-income countries, where dogs are the principal reservoir of rabies [1]. There are two ways to limit human rabies deaths: delivering timely post-exposure prophylaxis and interrupting the transmission of rabies virus in dogs, the reservoir host. The advantage of interrupting transmission is that it addresses the source of the problem and provides enormous, long-term public health and economic benefits. The World Health Organization (WHO) has set the goal of zero human deaths from dog-mediated rabies by 2030. The proposed mechanism for accomplishing this is through vaccinating at high enough coverage (≥70%) to achieve herd immunity.
With more than 20,000 human deaths each year due to dog-transmitted rabies, India has the highest disease burden [2, 3]. Furthermore, India also has the largest free-ranging dog population in the world (~59 million) [4]. Achieving and sustaining high vaccination coverage across large geographic scale is a daunting challenge due to high population turnover rates, reliance on community participation to access dogs, cost, and lack of political will [5]. At present, rabies and dog population control programs in India are limited to a few urban centers. Ironically, most dog-mediated human rabies deaths occur in rural areas of India [6].
Overarching Goal
The objective of this research is to find focused, efficient strategies for interrupting dog to dog transmission of rabies virus in resource-limited settings. This will make a major contribution to eliminating dog-mediated human rabies deaths in India and beyond.
Central Hypothesis
Our scientific hypothesis is that rabies arrives recurrently in rural dog populations through stepping-stone dispersal originating in urban populations where the disease is endemic (Figure 1). Because dog population size is large in urban areas and small in rural villages, we also hypothesize that stochasticity is important in persistence (or lack thereof).
References
[1] Cleaveland S, Kaare M, Knobel D, Laurenson MK. Canine vaccination-Providing broader benefits for disease control. Vet Microbiol. 2006;;117: 43–50.
[2] Burki T. The global fight against rabies. Lancet. 2008;;372: 1135–1136.
[3] Sudarshan MK, Madhusudana SN, Mahendra BJ, Rao NSN, Ashwath Narayana DH, Abdul Rahman S, et al. Assessing the burden of human rabies in India: results of a national multi-center epidemiological survey. Int J Infect Dis. 2007;;11: 29–35.
[4] Gompper ME. The dog-human-wildlife interface: assessing the scope of the problem. In: Gompper ME, editor. Free-Ranging Dogs and Wildlife Conservation. New York, USA: Oxford University Press;; 2014. pp. 9–54.
[5] Arechiga Ceballos N, Karunaratna D, Aguilar Setien A. Control of canine rabies in developing countries: key features and animal welfare implications. Rev Sci Tech l’OIE. 2014;;33: 311–321. doi:10.20506/rst.33.1.2278
[6] Suraweera W, Morris SK, Kumar R, Warrell DA, Warrell MJ, Jha P. Deaths from Symptomatically Identifiable Furious Rabies in India: A Nationally Representative Mortality Survey. PLoS Negl Trop Dis. 2012;;6. doi:10.1371/journal.pntd.0001847
Project Team: Craig Miller (Co-PI), Holly Wichman (Co-PI), JT Van Leuven, LuAnn Scott
Vaccines are a remarkably effective way to stem the threat posed by infectious diseases. Methods that allow rapid development of vaccines are vital. Synonymous recoding of viral genomes is a recently developed, general, and highly promising strategy for producing live attenuated vaccines. From an antigenic perspective, the method is ideal because it leaves the amino acid sequence of the viral proteins identical to the circulating pathogenic form. A number of viruses have been attenuated by recoding with non-preferred codons or codon pairs, and at least eight studies have shown protective immunization of mice. Despite its demonstrated success, there are fundamental gaps in our knowledge: 1) no effort has been made to compare alternative recoding strategies within the same virus in the same study; 2) several potential methods of synonymous recoding have not been tested at all; 3) the way in which attenuation is affected by the combination of multiple recoded genes is not known; and 4) most importantly, it is unresolved whether viruses attenuated by synonymous recoding are robust to evolutionary recovery.
This proposal tackles these gaps through three Specific Aims.
The project takes advantage of a bacteriophage model system with well-developed tools for genome manipulation and methods for rapid experimental evolution relative to eukaryotic viral systems (i.e., a hundred generations per day at very large population sizes). Achieving these three aims will yield approaches that can be applied to other systems for designing viruses with targeted levels of attenuation that are robust to evolutionary recovery. This research is a critical step toward the long-term goal of achieving a general strategy for fighting infectious diseases by precision design of live vaccines that do not re-evolve virulence when used in humans.